
In Silico Tools for Regenerative Medicine
In Silico Tools for Regenerative Medicine
One of the major challenges in improving medical devices and regenerative medicine strategies is understanding the exact interaction between the biomaterial and the human body. Our group strongly believes that an interdisciplinary approach which combines experimental with computational research is crucial to increase our fundamental knowledge, reduce the trial-and-error of experimental research and move towards more predictive cell-biomaterial interactions and improved medical devices and tissue engineered products. We focus on computational modelling of biological processes and cell-biomaterial interactions, using a range of data-driven to mechanistic modeling approaches, covering the intracellular and cellular scale as each in silico model system has its own benefits and limitations which determines the application for which it can be used. We are currently active in developing in silico models and machine learning algorithms to
- design advanced micropatterned and microfluidic high-throughput screening platforms,
- to gain fundamental understanding of cell-biomaterial interactions,
- to analyze high-throughput screening data and
- to inform the design and bioprocessing of regenerative medicine products,
as schematically shown in the figure on the right. Importantly, to calibrate and validate the in silico predictions we closely work together with colleagues of the MERLN Institute, the MDR program and REGMEDXB.
Selected publications
- King, J., Swapnasrita, S., Truckenmüller, R., Giselbrecht, S., Masereeuw, R., Carlier, A. (2021) Modeling indoxyl sulfate transport in a bioartificial kidney: two-step binding kinetics or lumped parameters for uremic toxin clearance? Computers in Biology and Medicine, 138: [104912], https://doi.org/10.1016/j.compbiomed.2021.104912
- Eroumé, K., Cavill, R., Stankova, K., de Boer, J., Carlier, A. (2021) Exploring the influence of cytosolic and membrane FAK activation on YAP/TAZ nuclear translocation. Biophysical Journal, 120(20): 4360-4377, https://doi.org/10.1016/j.bpj.2021.09.009
- Karagöz, Z., Geuens, T., LaPointe, V.L.S., van Griensven, M., Carlier, A. (2021) Win, lose or tie: mathematical modeling of ligand competition at the cell-extracellular matrix interface. Frontiers in Bioengineering and Biotechnology, section Tissue Engineering and Regenerative Medicine, 9: [657244], https://doi.org/10.3389/fbioe.2021.657244
- King, J., Eroumé, K., Truckenmüller, R., Giselbrecht S., Cowan, A.E., Loew, L., Carlier, A. (2021) 10 steps to investigate a cellular system with mathematical modeling. PLoS Computational Biology, 17(5): [e1008921], https://doi.org/10.1371/journal.pcbi.1008921
- Yang, L., Pijuan-Galito, S., Suk Rho, H., Vasilevich, A., Dede Eren, A., Ge, L., Habibovic, P., Alexander, M., de Boer, J., Carlier, A., van Rijn, P., Zhou, Q. (2021) High-throughput methods in the discovery and study of biomaterials and materiobiology. Chemical Reviews, 121(8), 4561–4677, https://doi.org/10.1021/acs.chemrev.0c00752
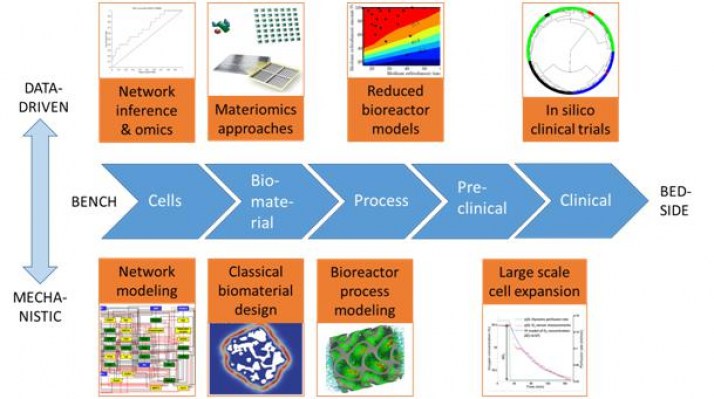 Schematic representation of the tissue engineering research and development process (horizontally) and the computer model classification (vertically). Figure is taken from: Geris, L., Lambrechts, T., Carlier, A., Papantoniou, I. (2018) The future is digital: in silico tissue engineering. Current Opinion in Biomedical Engeering,https://doi.org/10.1016/j.cobme.2018.04.001
Schematic representation of the tissue engineering research and development process (horizontally) and the computer model classification (vertically). Figure is taken from: Geris, L., Lambrechts, T., Carlier, A., Papantoniou, I. (2018) The future is digital: in silico tissue engineering. Current Opinion in Biomedical Engeering,https://doi.org/10.1016/j.cobme.2018.04.001







